 T22G5.7 vs. F56D2.2 T22G5.7 vs. F56D2.2 | 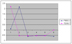 |
| Match score: 5.995 |
| T22G5.7 |
|
(predicted coding sequence) Saposin type B. Saposins are small lysosomal proteins that serve as activators of various lysosomal lipid-degrading
enzymes. They probably act by isolating the lipid substrate from the membrane surroundings, thus making it more accessible to the soluble degradative enzymes. A
ll mammalian saposins are synthesized as a single precursor molecule (prosaposin) which contains four Saposin-B domains, yielding the active saposins after prot
eolytic cleavage, and two Saposin-A domains that are removed in the activation reaction. The Saposin-B domains also occur in other proteins, many of them active
in the lysis of membranes. (From InterPro)
|
| F56D2.2 |
|
(predicted coding sequence) RING finger. The RING-finger is a specialized type of Zn-finger of 40 to 60 residues that binds two atoms of zinc, a
nd is probably involved in mediating protein-protein interactions. There are two different variants, the C3HC4-type and a C3H2C3-type, which is clearly related
despite the different cysteine/histidine pattern. The latter type is sometimes referred to as 'RING-H2 finger'. E3 ubiquitin-protein ligase activity is intrinsi
c to the RING domain of c-Cbl and is likely to be a general function of this domain; Various RING fingers exhibit binding to E2 ubiquitin-conjugating enzymes (U
bc's). (From InterPro)
|
| Description |
|
Our algorithm identified a time-delayed relationship between these two genes. F56D2.2 is expressed first and then T22G5.7. In the endocytosis pathway, some memb
rane proteins are ubiquitinated first by the ubiquitination enzymes including E3 ligase and then are sorted into lososomes and get degraded, which fits with the
relationship identified by us.
|
 |
 F02C12.5A vs. C34D1.5 F02C12.5A vs. C34D1.5 | 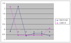 |
| Match score: 5.959 |
| F02C12.5A |
|
(predicted coding sequence) Cytochrome P450 enzyme. The cytochrome P450 enzymes usually act as terminal oxidases in multicomponent electron tr
ansfer chains, called P450-containing monooxygenase systems. P450-containing monooxygenase systems primarily fall into two major classes: bacterial/mitochondria
l (type I), and microsomal (type II). All P450 enzymes can be categorised into two main groups, the so-called B- and E-classes: P450 proteins of prokaryotic 3-c
omponent systems and fungal P450nor (CYP55) belong to the B-class; all other known P450 proteins from distinct systems are of the E-class. This family contains
a number of subtypes of both B and E classes. (From InterPro)
|
| C34D1.5 |
|
(predicted coding sequence) bZIP (Basic-leucine zipper) transcription factor family. The bZIP superfamily of eukaryotic DNA-binding transcriptio
n factors groups together proteins that contain a basic region mediating sequence-specific DNA-binding followed by a leucine zipper required for dimerization. (
From InterPro)
|
| Description |
|
bZIP transcription factors maybe activate the Cytochrome P450 enzyme transcription (not confirmed by the experiments). So bZIP transcription factors should be e
xpressed first and then the P450 enzyme is transcripted, which fits with the time-delayed relationship by our algorithm.
|
 |
 K03A1.5 vs. F26D10.10 K03A1.5 vs. F26D10.10 | 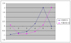 |
| Match score: 5.995 |
| K03A1.5 |
|
(confirmed gene) AMP-dependent synthetase and ligase. A number of prokaryotic and eukaryotic enzymes, which appear to act via an ATP-dependent c
ovalent binding of AMP to their substrate, share a region of sequence similarity. This region is a Ser/Thr/Gly-rich domain that is further characterised by a co
nserved Pro-Lys-Gly triplet. The family of enzymes includes luciferase, long chain fatty acid Co-A ligase, acetyl-CoA synthetase and various other closely-relat
ed synthetases. (From InterPro)
|
| F26D10.10 |
|
(predicted coding sequence) Glutamine synthetase. Glutamine synthetase (GS) plays an essential role in the metabolism of nitrogen by catalyzin
g the condensation of glutamate and ammonia to form glutamine. (From InterPro)
|
| Description |
|
Amino acids (such as glutamate) are made from intermediates of the citric acid cycle and other major pathways. And Acetyl-CoA is one of the most important subst
rate of citric acid cycle. Our algorithm has identified a time-delayed relationship between these two genes. K03A1.5 is expressed first and then F26D10.10 is ex
pressed, which fits perfectly with their biological functions.
|


T22G5.7 vs. F56D2.2
F02C12.5A vs. C34D1.5
K03A1.5 vs. F26D10.10


F31E8.6 vs. C34E10.7
